What are AAVs? and what makes them good candidates for gene therapy?
By: Stephen Malina, Eryney Marrogi, and Christopher Reardon.
In two previous posts, we introduced gene therapy, a method for curing genetic diseases by providing healthy copies of defective genes, and Adeno-associated virus (AAV) capsids, the gene therapy delivery system Dyno focuses on. In those posts, we also discussed how natural variants of AAV did not evolve for the specialized functions to which we now seek to apply them, which is why Dyno is applying machine learning and high-throughput techniques to better engineer AAV.
This post (the first in a three part series on AAV) provides an overview of AAV as a gene therapy vector, focusing primarily on the genetic and protein structure of an AAV capsid. Before diving in, we want to note upfront that this series is not intended to be a comprehensive overview of AAV research. Instead, it’s better thought of as an overview of selected AAV knowledge we at Dyno have learned and found helpful while working in this space. While all readers are welcome, in writing this post, we’ve tried to optimize for readers who may have a minimal biology background but no specific knowledge of virus biology and/or AAV but are eager to learn. In addition to summarizing AAV biology and engineering basics, we’ve given our perspective on some promising directions in AAV engineering and a sneak peek at Dyno’s AAV engineering workflow.
With that, let’s dive in.
AAV Vector Biology
Genome
Viruses evolved to infect cells and leverage their cellular machinery to make copies of themselves, and AAV is no exception. In order to accomplish this AAV first infects a cell, allowing its genetic material to get shuttled into the nucleus for transcription. Second, cellular machinery translates AAV’s viral genome into proteins which assemble into a viable viral shell with a copy of the viral genome packaged inside it. What distinguishes AAV from many other viruses is 1) its small genome size and 2) its inability to replicate itself in the absence of a helper virus, hence why the third step, “replicate exponentially and destroy the cell” is missing.
As a single-stranded DNA virus, AAV’s genome comprises 4.7 kb of linear DNA. This short DNA amazingly codes for at least 9 distinctly expressed proteins, which span a total unrolled length of over 12 kb. To achieve this, the genome also takes advantage of staggered and alternative start sites and splicing to create multiple proteins. In practice, this means that the same DNA nucleotides often simultaneously encode different segments of multiple proteins in the AAV genome. AAV’s small size is a double-edged sword from our perspective as AAV engineers. On one hand, its simplicity makes understanding its function more tractable. On the other hand, its heavy use of DNA-overlap-tricks makes engineering more difficult because a single nucleotide mutation can impact multiple proteins.
As mentioned, AAV lacks the full set of genes required to make copies of itself in a cell (it is replication incompetent). In addition to relying on the genetics and cellular machinery of its host organism, it needs the genes of the Adenovirus (hence the name adeno-associated virus) in order to replicate. This replication incompetence makes AAV useful for gene therapy. Gene therapy vectors are intended to deliver therapeutic genes without making copies of themselves. Whereas other vectors require careful engineering to handicap their ability to replicate in vivo, with AAV we get this property automatically.
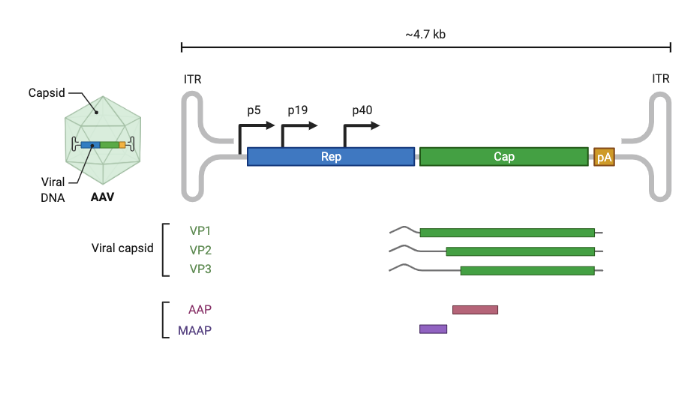
Amongst the genes that are involved in AAV’s functions, the two most critical are rep and cap. The rep gene encodes four proteins and plays a key role in enabling AAV replication. For the purpose of gene therapy, rep is only used during the AAV production stage. The cap gene, short for capsid, encodes three vector proteins VP1, VP2 and VP3, which combine to form the shell of the AAV capsid (see next section for additional details). In addition to its coding regions, the ends of the AAV ssDNA sequence are flanked by two inverted terminal repeats (ITRs) made up by repeated sequences that self-complement, thus allowing the structure to fold and provide stability to each end of the genome as a defense against degradation. The ITRs also play a key role in integration and rescue to and from the host cells, loading of the genome into the AAV capsid particle, and even act as promoters for second strand synthesis and protein expression (source).
When converting a natural AAV into a gene therapy vector, its genome is dissected, manipulated, and reassembled to make room to include therapeutic genes (transgenes). Research into the basic biology of AAV has enabled transformation of the natural AAV genome into a safe and effective gene therapy vector.
Structure
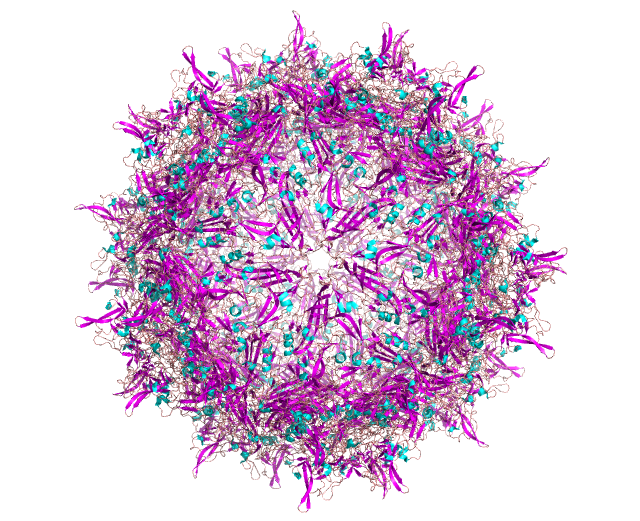
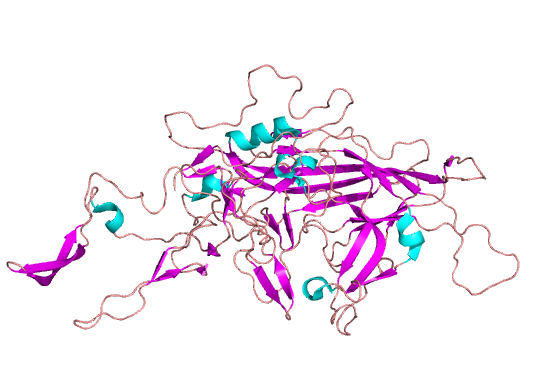
Model of AAV with all monomers assembled (left) and a single composite of the three AAV capsid monomers (VP1/2/3) (right), with coloration by secondary structure.
AAV’s prominence in gene therapy can also be understood by examining its structure (visually depicted above). Out of its two genes, the cap gene has a larger influence on the structure-function relationship. To start, cap expresses structural proteins VP1, VP2 and VP3, which interact to form the viral capsid from sixty copies of the VP monomers. Beyond contributing to the iconic icosahedral form, cap plays a critical role in determining tropism, the virus’ ability to infect a particular cell or organ type. Due to the capsid protein’s importance in capsid attachment to cellular receptors involved in tissue tropism, manipulation of the cap gene appears to be the best route to the selective tropism needed for gene therapy applications.
Function
AAV engineering focuses heavily on addressing the interaction of AAV with host cells in an attempt to manipulate viral entry, i.e. transduction. While each AAV serotype might interact with unique receptor proteins, all serotypes (natural variants) follow the same general mechanism (depicted diagrammatically below) for entering and transducing a cell. For example, one of the most studied serotypes, AAV2, identifies viable target cells through cellular receptor heparan sulfate proteoglycan (HSPG). Once the capsid finds a valid binding site, AAV2 enters the cell via cell-mediated endocytosis through clathrin-coated pits, and eventually escapes its endosome somewhere within the cytoplasm to attempt entry into the nucleus. Finally, the viral vector enters the nucleus through the nuclear pore complex, where transcription takes place [1].
After establishing itself within the nucleus, AAV relies on host-cell mechanisms for both genome replication and, in the case of engineered viruses, transgene expression (conversion into proteins). When designing capsids for therapeutic use, engineering the capsid can help the AAV find a target host cell, and engineering other components of the AAV can further improve the therapeutic. For example, AAV constructs can be selectively expressed within specific cell types by careful promoter expression, allowing increased control of engineered viruses in a therapeutic context. The right promoter, within the context of AAV, can dictate when and where an AAV construct starts expressing, ultimately contributing to making the therapy safe and effective. Promoter selection, coupled with capsid engineering are our main tools when making AAV a powerful tool for gene therapy.
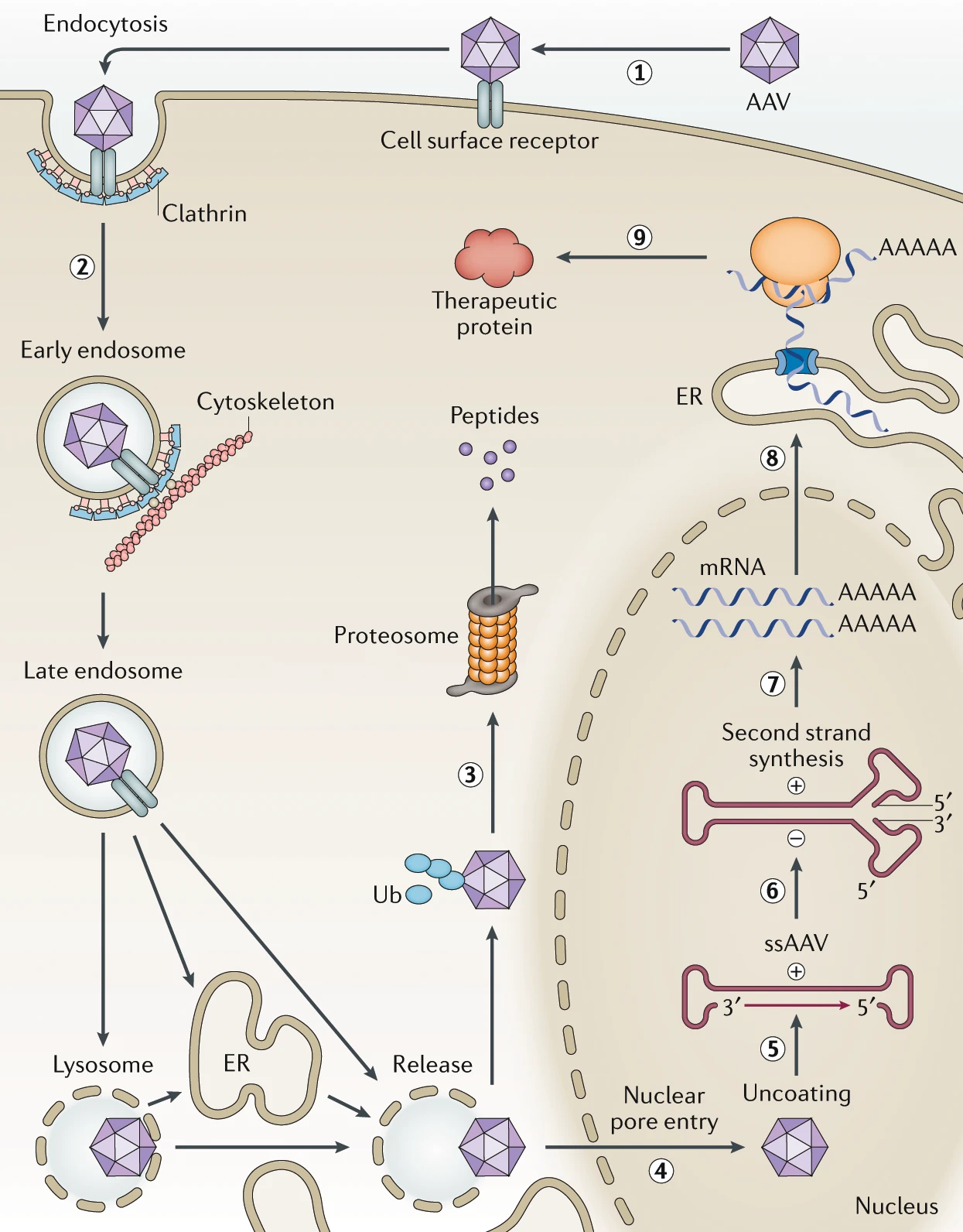
Source: Engineering adeno-associated virus vectors for gene therapy [2]
Conclusion
During its 50 year history, the field of AAV biology has built up an impressive edifice of knowledge about AAV’s genome, structure, function, and engineering. We’ve only scratched the surface of each of these topics in this post, but we have hopefully provided enough of an overview to help you understand the potential of engineered AAV as a gene therapy vector and some of the challenges the field needs to overcome in order to realize this potential.
Finally, this wouldn’t be a company blog post if we didn’t mention that we’re currently hiring for a range of technical and non-technical roles. If you’re excited about working with us, please apply and for the authors’ sake, mention that our blog post played a role in getting you excited about Dyno!
Special thanks to: Adam Poulin-Kerstein, Alex Brown, Cherry Gao, Eric Kelsic, Heikki Turunen, Jeff Gerold, and Sam Sinai for helpful comments.
Bibliography
[1]: Martini, S. V., P. R. M. Rocco, and M. M. Morales. “Adeno-associated virus for cystic fibrosis gene therapy.” Brazilian Journal of Medical and Biological Research 44 (2011): 1097-1104.
[2]: Li, C., Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet 21, 255–272 (2020). https://doi.org/10.1038/s41576-019-0205-4