By: Sam Sinai
As 2024 comes to a close, I look back at one of Dyno’s most exciting technical developments. Earlier this year, Dyno introduced LEAPSM: Low-shot Efficient Accelerated Performance. With as few as 19 designs in one experimental batch, LEAP can achieve performance improvement equivalent to what previously required at least an additional round of high-throughput in vivo experiment (animal testing). Moreover, LEAP is approaching a success rate in designing high-functioning proteins where it would be feasible to directly test its computational designs in single-candidate validation experiments. This future would dramatically cut the time needed for therapeutic development and eventually reduce the cost of treatment for patients.
The Promise of Machine Learning in Therapeutics
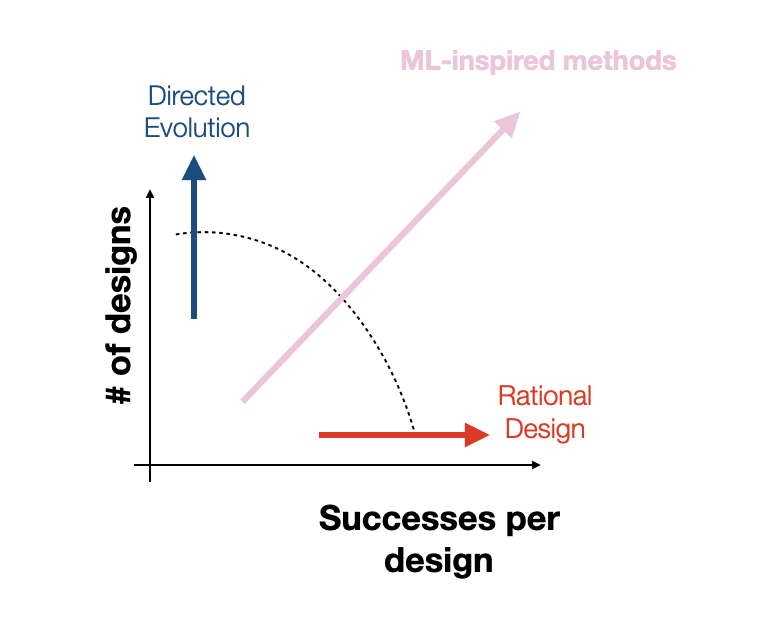
A key promise of machine learning (ML) in biology is to relieve researchers of the burden of laborious and costly experiments, empowering them to solve higher leverage problems. Designing proteins and testing their efficacy in vivo typically demands multiple rounds of complex trials. Dyno (and the field) hopes that ML can cut through some of this, discerning the underlying principles of protein design and generating sequences with desired functionalities. We want therapeutics that translate to in vivo settings, i.e. that work in humans, and we want to find them fast. It is important to note that the real bottleneck for speed is not how many good looking sequences you can propose in silico (purely computationally), but how many of the ones you test will actually translate to real treatments. With LEAP, we were able to discover high performing AAVs in silico, with a very high per-attempt translation rate in non-human primates, effectively cutting out the need for at least one high-scale animal experiment.
The Challenge of Designing Complex Proteins like AAVs
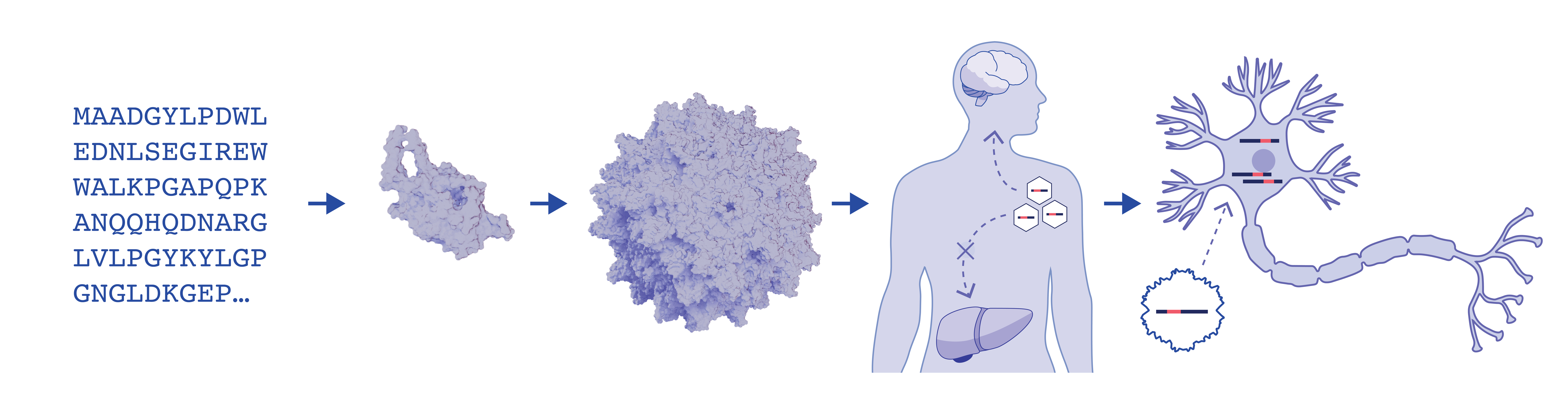
In this post, I show a case study on Dyno’s favorite protein: The Adeno-Associated Virus (AAV) capsid. The AAV capsid is a complex protein with structural and enzymatic parts that needs to fold, assemble, package a genome, evade the body’s defenses, enter specific cells and deliver its DNA cargo in the nucleus in order for a therapy to be effective. A capsid should also avoid entering off-target organs such as the liver, where it can cause toxicity. The success or failures of these functions is determined by the protein sequence of the capsid. Effective delivery of genes into specific organs and cells has been a challenging bottleneck of gene therapy.
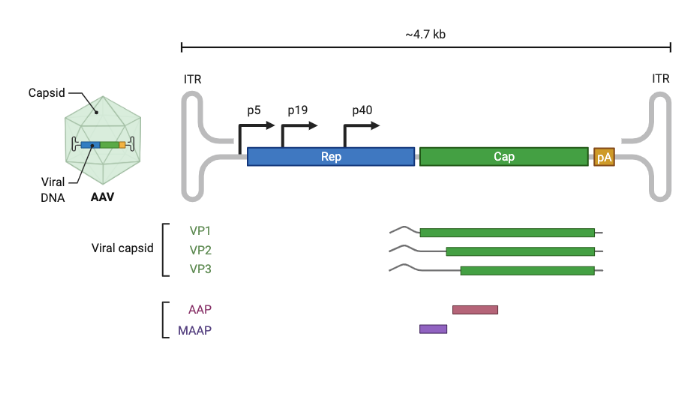
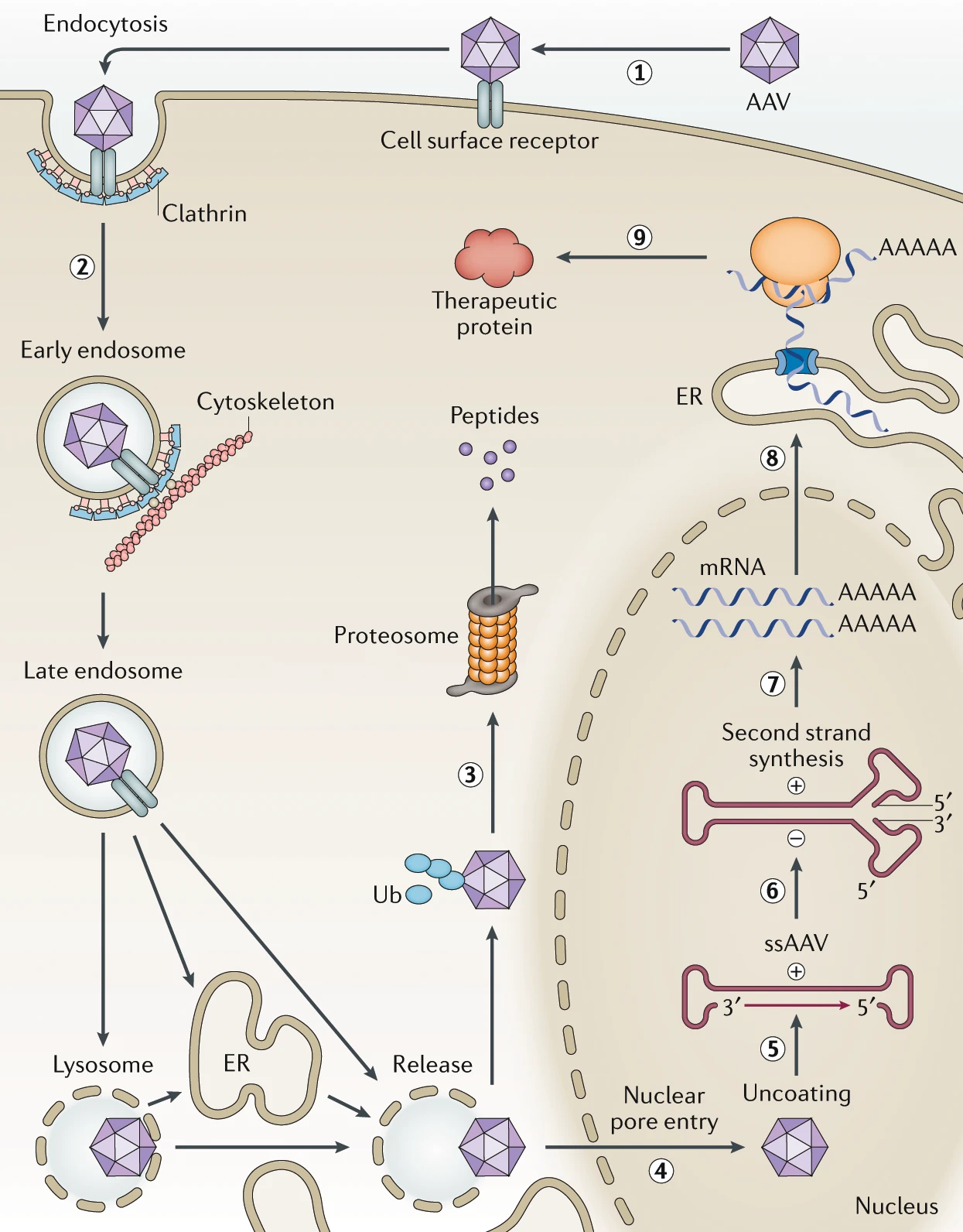
The anatomy of a 60 year problem. A ~735 amino-acid chain is the building block for the 60-meric AAV capsid. This sequence determines whether the intravenously-administered capsid can be manufactured, avoid neutralization and off-target transduction, cross the blood-brain barrier, and deliver its cargo to neurons.
Designing capsids with specific traits—such as efficient targeting of brain cells and avoidance of liver tissue—requires predicting the success of each step of the process depicted above, either through mechanistic understanding of the biology (“white-box”) or by employing ML methods trained on experimental endpoints (“black-box”; prediction without explicit knowledge of the mechanism). Despite years of progress, a comprehensive mechanistic picture for AAVs remains distant. Importantly, understanding the mechanisms that enable a particular sequence to perform well does not guarantee insights into how another sequence might achieve the same (or better) phenotypic outcomes through different means.
On the machine learning side, the capsid’s behavior is poorly predicted by zero-shot metrics extracted from structure-based and protein language models. Generally speaking these methods are far from good proxies for predicting function for complex proteins, and even for simpler in vitro (in lab media, but not in organisms) experiments tend to have low accuracy as the designs diverge from natural protein sequences.
Dyno adopts a “grey-box” approach to designing AAV capsids with novel properties. While primarily relying on black-box models and empirical data, we incorporate mechanistic knowledge through models and experiments when available.
Current Approach to AAV Discovery
Consider a typical process of in vivo capsid discovery. Usually, 10,000 to 10 million pooled sequences are screened in mice or non-human primates (NHP) to identify the best-performing capsids in terms of tissue targeting. This high-throughput discovery screen is then usually repeated, using insights from previous rounds to refine the sequence design. Then, a smaller subset of 10 to 100 sequences is selected for a follow-up validation experiment, which typically leads to a final group of 1 to 5 candidates that undergo single-capsid testing in primates. This final stage—involving histological analysis necessary for clinical trial clearance—is expensive and reserved for only the most promising candidates. For in vivo AAV studies, each experiment could cost upwards of a million dollars and can take nine months to complete.
LEAP: Pushing the Boundary of Possibility with Many Models, but Few Attempts
In LEAP, we train a mixture of tens of partially independent and calibrated models: predictors, filters, and generators, to propose and sift through a large set of virtual sequences. Models range from those pre-trained on public data, to fine-tuned or trained from scratch on different slices or attributes of Dyno’s internal data. Our generative methods can propose a high performing, diverse set of candidates and our filtering ensembles can accurately eliminate sequences that are unlikely to be extremely good. Designing capsids that are much better than anything you have trained on is hard to get right. As articulated well here:
“When designing objects to achieve novel property values with machine learning, one faces a fundamental challenge: how to push past the frontier of current knowledge, distilled from the training data into the model, in a manner that rationally controls the risk of failure. If one trusts learned models too much in extrapolation, one is likely to design rubbish. In contrast, if one does not extrapolate, one cannot find novelty” – Fannjiang & Listgarten, 2023
To use an analogy, it is as if you train an AI system on tweets, all with less than 1000 likes, and ask it to propose a tweet that is liked 10,000 times, with as few as 10 attempts. This type of “out-of-distribution” extrapolation, finding high performance (often across multiple dimensions) you do not observe in your training data, is the most critical dimension in therapeutics design, and importantly current frontier models are relatively weak at optimizing in this dimension.
Because of our confidence in the LEAP’s proposals, Dyno can bypass earlier rounds of experimental de-risking and directly test designs in high-stakes experiments. This is replacing years of experiments with days of in silico design and compute.
Results in the Primate Brain
I show the results of one of our first campaigns using LEAP below. In this experiment, we deployed LEAP to design capsids that target the brain. This was a medium throughput validation experiment (10s of unique capsids measured together in each animal) that measures in vivo performance of capsids with high resolution (including cell-type specificity). Our measurement endpoints are:
- Packaging: whether the capsid successfully assembles and packages its genome.
- Transduction: whether we detect a higher frequency of transduction events, measured (with careful statistical and experimental controls) through mRNA readouts within the brain.
- De-targeting: whether we detect lower abundance of viral DNA in the liver, relevant for the safety profile.
Due to the high per-design cost of these experimental rounds (e.g. each capsid is independently manufactured by our lab), our standard approach has been to test variants that have performed really well in at least one previous high-throughput (100K-1M designs) experiment. We also task our in-house protein experts to modify those variants in small but rational ways to improve their targeting. The most risky bet is to just design variants we never observed before with ML and test them in such high labor experiments directly. That’s exactly what we did. We allocated 19 LEAP-designed capsids, which we had never measured before, along with our standard approach (all designed on the AAV9 backbone). The capsids designed by LEAP were at least four and often 7-10 (non-contiguous) mutations away from any samples measured in the training set (i.e. virtually impossible to find either at random or rationally). The outcome was striking: not only were a majority of our LEAP designs functional, but about half of them improved on the best known design. The best of these designs improved as much over the previous round as can be expected from a successful high throughput or directed evolution round. It is also noteworthy that at design time, there is some uncertainty about which high-throughput measured variant will be the absolute best in follow up (the 1x benchmark), and therefore, LEAP designs are derived from a collection of diverse sequences, rather than many modifications to the benchmark we compare against.
LEAP’s performance in the NHP (cyno) brain. LEAP-designed capsids (red) compared to top capsids drawn from a previous discovery round (grey) and rationally improved capsids (purple). (Upper) Empirical Cumulative distribution function (CDF) showing the proportion of capsids from each category with brain transduction rates greater than the value indicated on the horizontal axis. Green shaded region indicates brain transduction values higher than any capsid drawn from the previous discovery round. (Lower) Scatter plot showing brain transduction and liver detargeting rates for each successfully packaged capsid.
To summarize:
- 17 out of 19, of the LEAP-designed capsids packaged successfully. This is a very high packaging rate considering that most single mutations to capsids break packaging (the very first property needed for every downstream task).
- 9 out of 19 LEAP capsids outperformed any previously observed sequence in terms of brain transduction, and all expert designs. Most of these also show more liver de-targeting, potentially improving the safety of a future therapy.
- Overall the brain transduction improvement compared to the previous best design was 6 fold, while also achieving much better liver detargeting. Comparatively, typical directed evolution or standard high throughput screens for capsid optimization achieve 2-5 fold improvement in a single round, but with sequence budgets up to 10M samples.
While I only discuss the result of one experiment, we’ve found LEAP to be consistently high-yield and high-performance across multiple trials. With about 50% of variants improving on the best sample in the training set, with as few as 5 designs, one can already expect (with high probability) to find at least 1 design that improves on anything that was observed before.
Today operating LEAP still requires careful supervision by ML experts, and the number of candidates we can evaluate in silico by our large ensemble of models (some slow at inference) is in the millions. A major direction of improvement is to enable large scale in silico screens, which would be enabled by higher compute deployment and better inference. As we make progress there, we are also working to reduce the expertise needed to operate a LEAP-enabled campaign.
Will LEAP work for your biological sequence?
Fundamentally, most techniques we use within LEAP are not specific to AAVs. While some models take advantage of the large datasets we’ve collected within Dyno, many are trained on smaller or public datasets. Performance quickly improves with more data, but our approach is robust to low-data regimes (though it is not a zero-shot system, i.e it requires at least some data in the relevant domain). In short, we expect LEAP to generalize for other biological sequences. This is why we are excited about our ML technology and its future impact on gene therapy and other relevant therapeutic domains.
In the coming year, we are exploring partnering with a limited number of companies to apply the techniques we’ve developed with LEAP to help solve in vivo design problems. If you are curious whether LEAP could be applicable to your own sequence design problems, I encourage you to get in touch (info@dynotx.com).
Special thanks to: David Brookes, Abhishaike Mahajan, Stephen Malina, Alice Tirard and Eric Kelsic for helpful comments on this post.